-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaIs a Novel Locus for Waist Circumference: A Genome-Wide Association Study from the CHARGE Consortium
Central abdominal fat is a strong risk factor for diabetes and cardiovascular disease. To identify common variants influencing central abdominal fat, we conducted a two-stage genome-wide association analysis for waist circumference (WC). In total, three loci reached genome-wide significance. In stage 1, 31,373 individuals of Caucasian descent from eight cohort studies confirmed the role of FTO and MC4R and identified one novel locus associated with WC in the neurexin 3 gene [NRXN3 (rs10146997, p = 6.4×10−7)]. The association with NRXN3 was confirmed in stage 2 by combining stage 1 results with those from 38,641 participants in the GIANT consortium (p = 0.009 in GIANT only, p = 5.3×10−8 for combined analysis, n = 70,014). Mean WC increase per copy of the G allele was 0.0498 z-score units (0.65 cm). This SNP was also associated with body mass index (BMI) [p = 7.4×10−6, 0.024 z-score units (0.10 kg/m2) per copy of the G allele] and the risk of obesity (odds ratio 1.13, 95% CI 1.07–1.19; p = 3.2×10−5 per copy of the G allele). The NRXN3 gene has been previously implicated in addiction and reward behavior, lending further evidence that common forms of obesity may be a central nervous system-mediated disorder. Our findings establish that common variants in NRXN3 are associated with WC, BMI, and obesity.
Published in the journal: . PLoS Genet 5(6): e32767. doi:10.1371/journal.pgen.1000539
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1000539Summary
Central abdominal fat is a strong risk factor for diabetes and cardiovascular disease. To identify common variants influencing central abdominal fat, we conducted a two-stage genome-wide association analysis for waist circumference (WC). In total, three loci reached genome-wide significance. In stage 1, 31,373 individuals of Caucasian descent from eight cohort studies confirmed the role of FTO and MC4R and identified one novel locus associated with WC in the neurexin 3 gene [NRXN3 (rs10146997, p = 6.4×10−7)]. The association with NRXN3 was confirmed in stage 2 by combining stage 1 results with those from 38,641 participants in the GIANT consortium (p = 0.009 in GIANT only, p = 5.3×10−8 for combined analysis, n = 70,014). Mean WC increase per copy of the G allele was 0.0498 z-score units (0.65 cm). This SNP was also associated with body mass index (BMI) [p = 7.4×10−6, 0.024 z-score units (0.10 kg/m2) per copy of the G allele] and the risk of obesity (odds ratio 1.13, 95% CI 1.07–1.19; p = 3.2×10−5 per copy of the G allele). The NRXN3 gene has been previously implicated in addiction and reward behavior, lending further evidence that common forms of obesity may be a central nervous system-mediated disorder. Our findings establish that common variants in NRXN3 are associated with WC, BMI, and obesity.
Introduction
Body mass index (BMI) is a commonly used measure of overall adiposity. However, specific fat depots may confer differential metabolic risk. In particular, central abdominal fat, as measured by waist circumference (WC), may be more strongly associated with the development of metabolic risk factors and cardiovascular disease as compared with BMI [1]–[4]. Therefore, understanding the pathogenesis of central fat distribution may provide further insight into the relationship between adiposity, cardiometabolic risk, and cardiovascular disease.
Both genetic and environmental factors have been linked to obesity [5]. Heritability estimates for BMI and WC range from 30 to 70% in family and twin studies [6], and multiple quantitative trait loci and candidate genes have been mapped to genes for central adiposity [5]. Despite strong evidence for an underlying genetic component, genes for obesity-related traits, particularly central obesity, have been difficult to identify and replicate.
Early genome-wide association studies (GWAS) identified both FTO and MC4R as genes related to BMI and WC [7]–[10]. Many new loci have been identified in recent obesity related GWAS studies [11]–[13]. However, collectively these variants explain only a small proportion of the variation in adiposity [7]–[13]. In addition, no GWAS exist exclusively to identify genes for central fat. Thus, to identify new variants, we carried out a large-scale meta-analysis of GWAS from eight studies to detect variants associated with central body fat distribution.
Methods
Study Samples
Participants for the current analysis were drawn from 8 cohort studies, including the Age, Gene/Environment Susceptibility-Reykjavik Study (AGES - Reykjavik Study), the Atherosclerosis Risk in Communities Study (ARIC), the Cardiovascular Health Study (CHS), the European Special Population Network consortium (EUROSPAN), the Family Heart Study, the Framingham Heart Study, Old Order Amish (OOA), and the Rotterdam Study (RS). These groups comprise the CHARGE (Cohorts for Heart and Aging Research in Genome Epidemiology) Consortium. All participants provided informed consent. Local ethical committees at each institution approved the individual study protocols. Text S1 contains details regarding all participating cohorts.
Imputation and Statistical Analysis
Common to all analyses were use of the raw WC measures and the assumption of an additive model; study specific details follow. Each study reported an effect allele which was meta-analyzed consistently across all studies. Results are currently presented relative to the minor G allele for the NRXN3 SNP. In all studies except CHS, MACH (version 1.0.15 in Family Heart, Framingham, EUROSPAN and RS; version 1.0.16 in ARIC, AGES, and OOA) was used to impute all autosomal SNPs on the HapMap, using the publicly available phased haplotypes (release 22, build 36, CEU population) as a reference panel. In CHS, the program BIMBAM was used [14]. Details are provided in Table S1 regarding covariates and trait creation.
In ARIC, Framingham, and RS, sex - and either cohort-specific or study center-specific residuals were created after adjustment for age, age-squared, and smoking status. In CHS and Family Heart, linear regression models were used to adjust for age, age-squared, sex, smoking, and study center. In AGES, linear regression models using PLINK v1.04 [15] were used to adjust for age, age-squared, sex, and smoking. In the OOA the measured genotype mixed effects model was used adjusting for age, age-squared, sex and family structure based on the complete 14-generation pedigree as implemented in ITSNBN [16]. Framingham employed the linear mixed effect model for continuous traits and the generalized estimating equations for dichotomous traits in R [17] to account for family relatedness. In RS, linear regression models were run using MACH2QTL [18]. In ARIC and EUROSPAN, all regression models were run using the ProbABEL package from the ABEL set of programs [19] and in EUROSPAN genomic control [20] was used to correct standard errors of the effect estimates for relatedness among individuals. The Family Heart Study determined the effect of each SNP using linear mixed effects models to account for the siblings present in the data using SAS.
Principal components calculated using EIGENSTRAT [21] were adjusted for in the individual studies when significant in order to account for population substructure.
Meta-Analysis
A weighted z-score approach was used to conduct meta-analyses with METAL (www.sph.umich.edu/csg/abecasis/metal/). Genomic control correction was applied to each study prior to the full meta-analysis. P-values less than 4.4×10−7 were considered genome-wide significant [22].
In Silico Exchange with the GIANT Consortium
In stage 2 of our study, we conducted an in silico exchange of the results of 48 SNPs with the GIANT consortium. To create our list of SNPs to exchange, we first selected the top 34 SNPs from independent loci (defined as SNPs with R2<0.2) from our meta-analysis of WC, excluding SNPs in known loci for adiposity. An additional 14 SNPs of independent loci with a p-value<1.0×10−5 from a secondary list that focused on SNPs for WC with corresponding BMI p-values>0.01 were also included in an attempt to isolate genes that might be specifically associated with central fat deposition. Our a priori threshold for replication was a p-value<0.001 (0.05/48 SNPs) and/or reaching genome-wide significance in a combined meta-analysis. CHARGE and GIANT results were then meta-analyzed using METAL.
Results
Table 1 presents descriptive statistics across the 8 cohorts providing data for the meta-analysis. We had a total sample size of 31,373 individuals of Caucasian descent. Participants were mostly middle-aged with ages ranging from a mean of 45 to 76 years of age.
Tab. 1. Descriptive statistics across the eight cohorts. 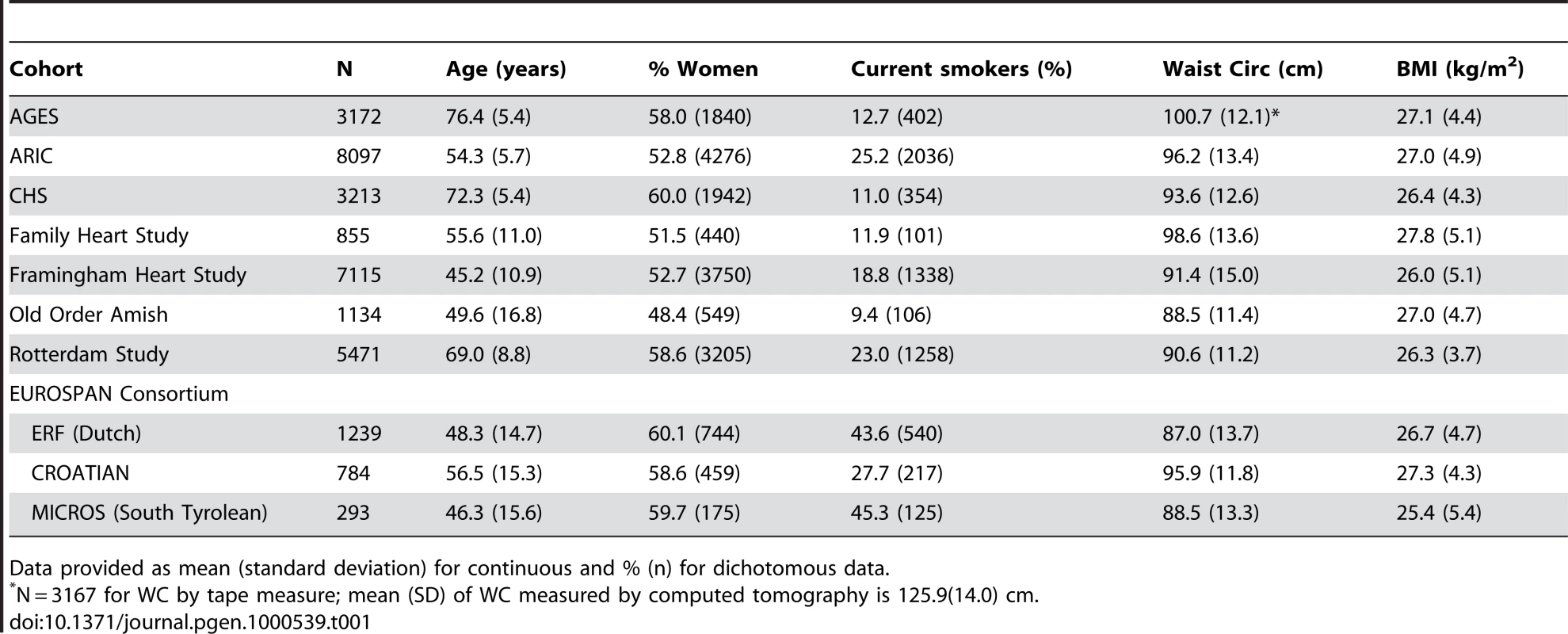
Data provided as mean (standard deviation) for continuous and % (n) for dichotomous data. Figure S1 shows the genome-wide association results for WC in the stage 1 CHARGE-only analysis. The top SNPs for WC were in the FTO and MC4R genes (Table S3). Figure S2 shows the QQ plot for our results excluding SNPs in FTO and MC4R. For FTO, the top SNP was rs1558902 (p = 4.6×10−19). For MC4R, the top SNP was rs489693 (p = 3.5×10−7). The top results excluding SNPs in FTO and MC4R from our stage 1 meta-analysis are shown in Table 2 along with the stage 2 in silico replication results from the GIANT consortium; additional meta-analysis results from CHARGE are presented in Table S3. The lowest p-value on our list, for SNP rs10146997 in the NRXN3 gene, had a stage 1 meta-analysis p-value of 6.4×10−7 and was confirmed in 38,641 participants from the GIANT consortium with a p-value of 0.009 and a combined p-value of 5.3×10−8. The NRXN3 SNP was derived from the list of SNPs associated with WC irrespective of association with BMI. None of the other SNPs that were exchanged were confirmed in GIANT. We do note that while rs10857809 (proxy for rs10857810) in the FAM40A gene had a p-value of 0.003 in GIANT, the results were not direction-consistent with CHARGE and therefore did not replicate in the combined analysis.
Tab. 2. Top 48 SNPs exchanged with the GIANT Consortium, GIANT p-values, and the combined results. 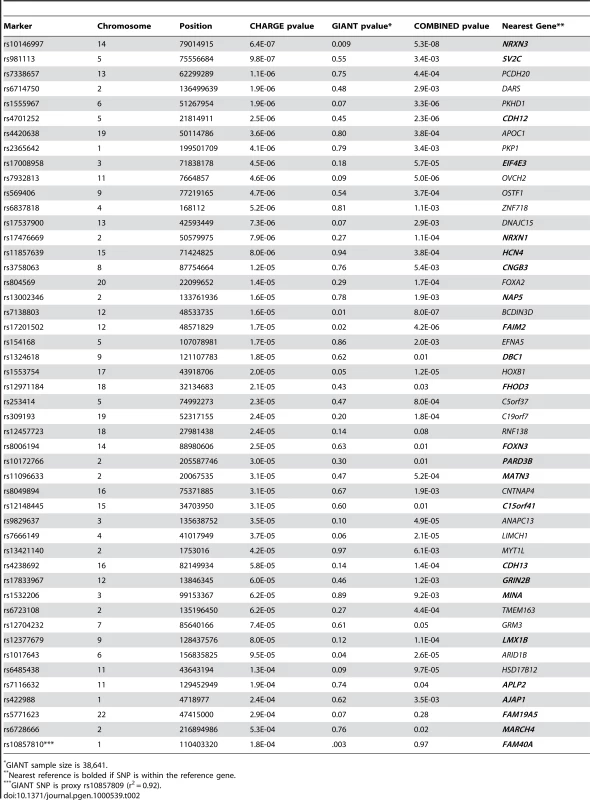
GIANT sample size is 38,641. Figure 1 presents the genomic region for SNP rs10146997 (intronic) in NRXN3. Table 3 shows detailed results of rs10146997 in the NRXN3 gene by contributing CHARGE study and corresponding results appear in the forest plot in Figure S3; there was no evidence for heterogeneity across the stage 1 studies (p = 0.64). The minor allele (G) frequency (MAF) for rs10146997 in our sample ranged from 0.14 in the OOA to 0.24 in the Croatians; the frequency of the NRXN3 SNP G allele is 0.275, 1.0, 1.0, and 0.35, in Hapmap CEPH, Han Chinese, Japanese, and Yoruba populations, respectively. This SNP was genotyped in AGES, CHS, Family Heart Study, Rotterdam and all EUROSPAN studies, and imputation scores for the other studies indicated very high quality. Overall, per copy of the G allele, mean WC was increased 0.0498 z-score units (0.65 cm). Beta coefficients (in z-score units) were consistently positive in all samples except the ERF study (β = −0.0098; p = 0.86), which is most likely due to chance. Due to overlap in participants from the Framingham Heart Study and ARIC with those from the Family Heart Study, the CHARGE meta-analysis was re-run for the NRXN3 SNP without the Family Heart Study; results were essentially unchanged (p = 6.6×10−7). Individual study-specific results for rs10146997 from the studies comprising the GIANT consortium can be found in Table S2.
Fig. 1. Regional Association Plot for rs10146997 on chromosome 14 in the stage 1 CHARGE-only analysis. 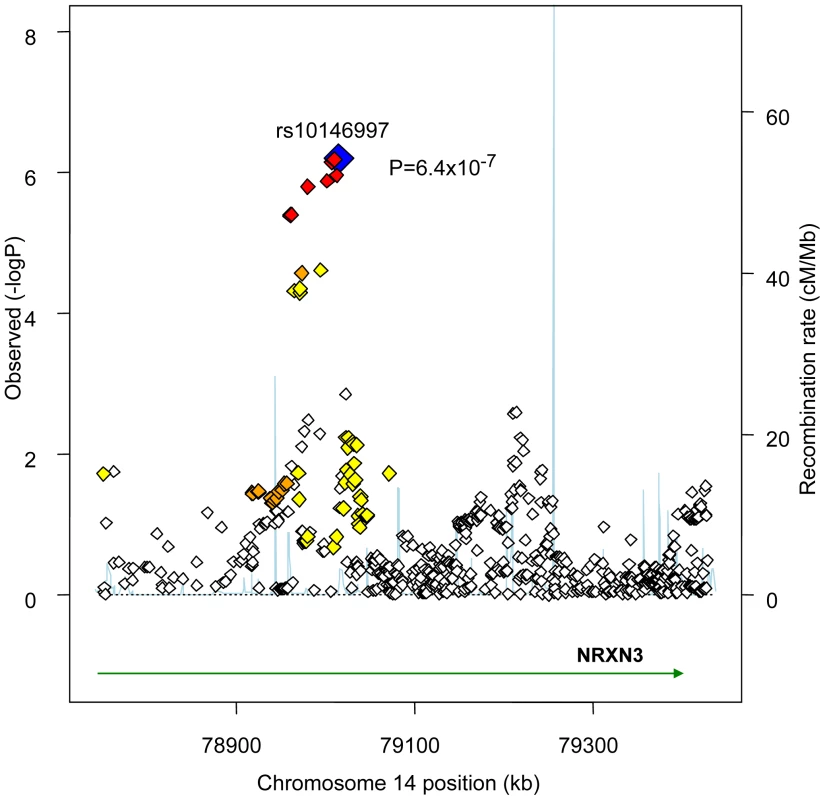
The color scheme is red for strong linkage disequilibrium (LD; r2≥0.8), orange for moderate LD (r2≥0.5 and <0.8), yellow for weak LD (r2≥0.2 and <0.5) and white for limited or no LD (r2<0.2). Tab. 3. Results per copy of the G allele for rs10146997 by contributing study; beta coefficients expressed as z-scores. 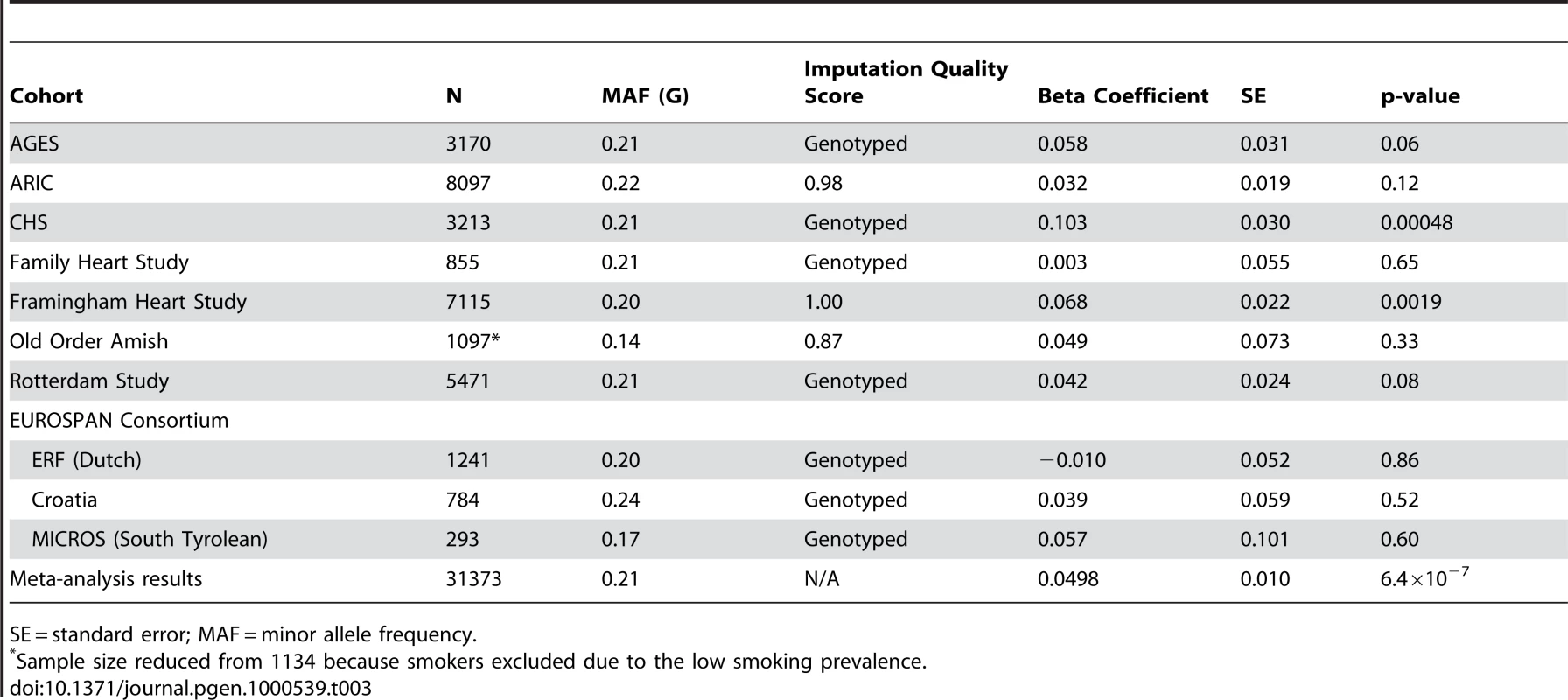
SE = standard error; MAF = minor allele frequency. Within CHARGE we also observed an association of rs10146997 with BMI (p = 7.4×10−6). Overall, mean BMI was increased 0.024 z-score units per G allele (0.10 kg/m2). When WC was additionally adjusted for BMI, the signal was completely attenuated (0.0065 z-score units per G allele; p = 0.32). The association of rs10146997 with WC was similar in women and men and in older and younger individuals (Table 4). After excluding smoking from the covariate adjustment list, results were essentially similar. Per copy of the G allele, the odds ratio of having high WC (≥88 cm in women; ≥102 cm in men) was 1.07 (95% CI 1.02–1.11; Table 4). Similarly, the odds ratio of obesity was 1.13 (95% CI 1.07–1.19).
Tab. 4. CHARGE consortium secondary analysis results per copy of the G allele for rs10146997 in 31373 individuals; beta coefficients expressed as z-scores. 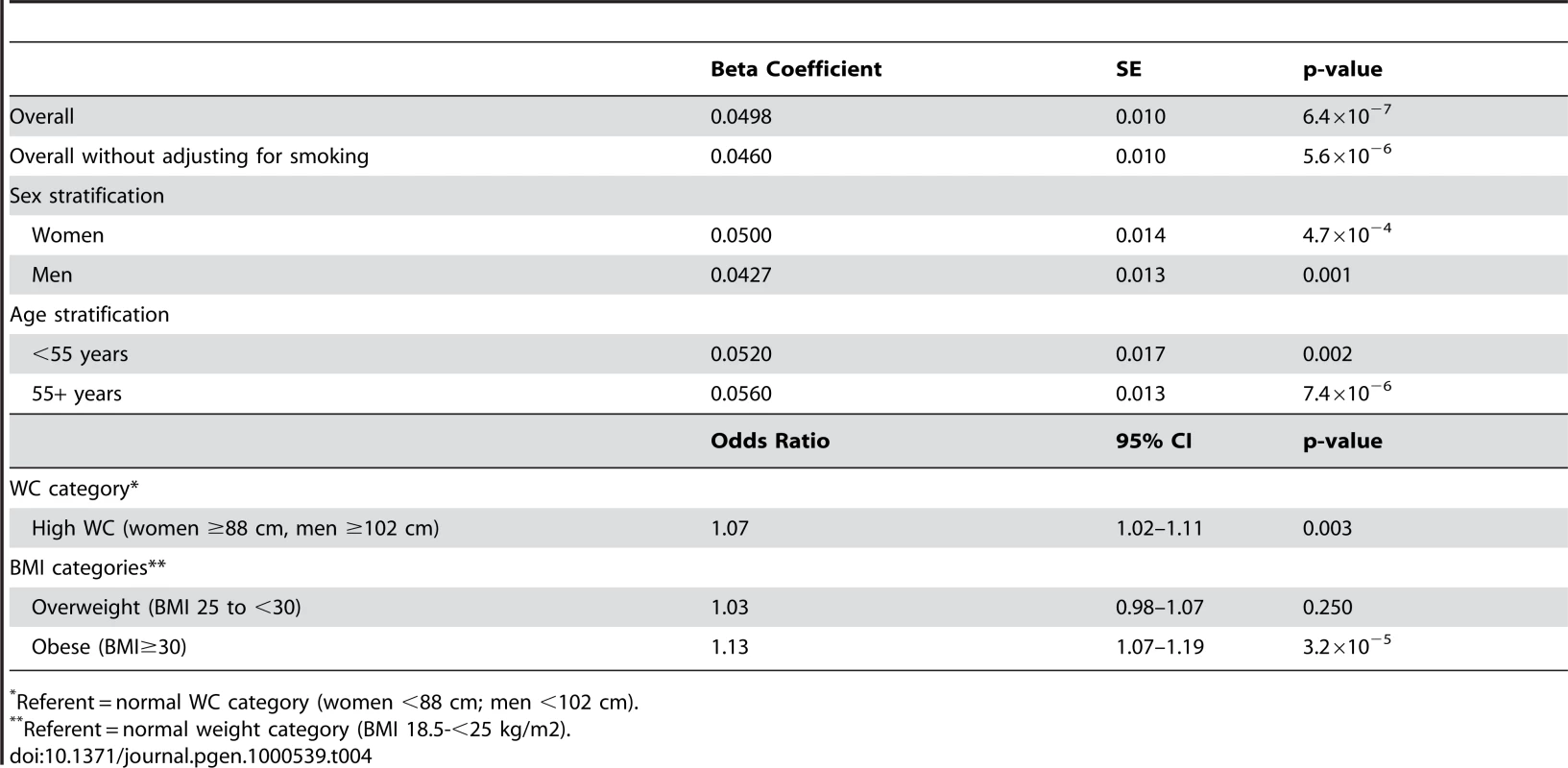
Referent = normal WC category (women <88 cm; men <102 cm). We calculated a risk score of FTO (rs9939609), MC4R (rs17782313), and NRXN3 with possible scores ranging from 0–6 risk alleles (Figure 2). Across this range, mean WC increased from 92.4 cm among those with 0 risk alleles, to 95.7 cm among those with 4 or more risk alleles. To put our findings in perspective, per copy of the effect allele, the NRXN3 SNP resulted in a WC difference of 0.65 cm; FTO 0.73 cm, and MC4R 0.37 cm.
Fig. 2. Mean waist circumference by number of risk alleles for FTO, MC4R, and NRXN3. 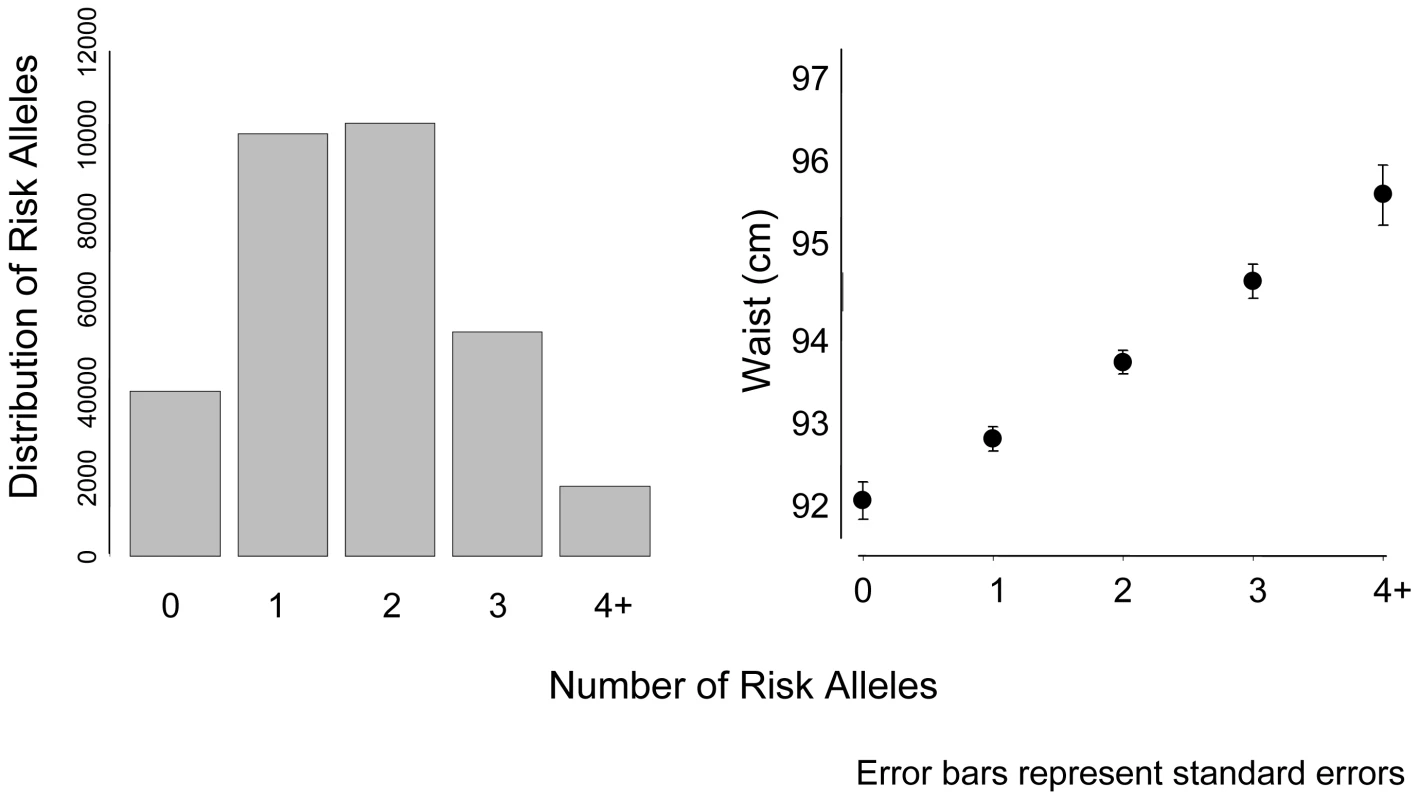
Bars represent standard errors. The panel on the left represents the distribution of risk alleles in the overall sample. CHARGE consortium meta-analysis results for BMI can be found in Table S4; Manhattan and QQ plots for BMI can be found in Figure S4 and Figure S5, respectively.
Discussion
In a discovery sample of more than 30,000 individuals from several cohort studies, we identified a novel locus in the NRXN3 gene associated with WC. In combination with data from the GIANT consortium, the p-value for this finding exceeded our pre-defined threshold for genome-wide statistical significance. This SNP was also significantly associated with BMI and obesity. This gene has previously been associated with addiction and reward behavior, and is a compelling biologic candidate for obesity. We also confirmed the significant associations with FTO and MC4R that have previously been reported.
Although our genome-wide scan was performed for WC, the NRXN3 SNP was also significantly associated with BMI. In secondary analyses, the signal for WC was attenuated after additionally adjusting for BMI, suggesting that this locus is most likely involved in overall adiposity and not specific to central fat deposition. Similar observations have been made for FTO [10] and MC4R [7], highlighting the inter-dependence between different measures of adiposity and the importance of performing GWAS on multiple adiposity-related traits.
The small magnitude of the effect size of the NRXN3 variant on WC is consistent with what has previously been reported for FTO and MC4R. These findings highlight the need for large sample sizes in order to facilitate continued gene discovery for obesity-related traits. In particular, genes that emerge for waist circumference will most likely be genes for overall adiposity because of the strong correlation between the two measurements [22]. More specific measures of visceral abdominal fat depots may make it possible to isolate genes involved in regional body composition.
NRXN3 is part of a family of central nervous adhesion molecules and is highly expressed in the central nervous system. Prior studies of NRNX3 point towards an important role in alcohol dependence, cocaine addiction, and illegal substance abuse [23]–[26]. In addition, opioid dependence has been linked to the chromosome 14q region [23]. In mice, NRXN3 beta expression was observed in the globus pallidus when exposed to cocaine [24]. Many of the neuronal pathways in these sub-cortical regions of the brain in which NRXN3 is expressed are involved with learning and reward training [25].
Obesity and addiction may share common neurologic underpinnings [26]. Other well-replicated obesity loci, including MC4R, have also been shown to be associated with centrally-mediated phenomena including binge eating behavior [11],[12],[27]. Studies in mice indicate that FTO expression is particularly pronounced in regions of the brain known to regulate energy balance [28], and recent data suggest that variants in the FTO gene may regulate food intake and selection [29].
Additional research is needed to understand the association of rs10146997 with the NRXN3 gene and to identify a causal variant. Since there are no other genes within a distance of more than several hundred kilobases of this SNP, it is unlikely that a different gene accounts for this finding. A search of publically available databases [30]–[32] did not identify an association between SNPs in NRXN3 and gene expression.
A relationship between WC and causal variants in the NRXN3 gene may have clinical implications. Obesity is a multifactorial trait that results from a complex interaction between genes and environment. The identification of an association between obesity and variants in a gene that has been associated with substance abuse suggests that further exploration of the role of this gene in vulnerability to addiction to food substances should be undertaken.
The strengths of this work include the large discovery sample size. The effect size was small, and achieving conventional levels of genome-wide significance required combining data from more than 70,000 participants in two large consortia. Although the confirmation with the GIANT consortium is promising, the joint p-value based on more than 70,000 participants achieved only borderline genome-wide significance. Our findings warrant the need for further replication in other ethnic groups.
We identified a SNP at a novel locus in the NRXN3 gene associated with WC. This gene has previously been implicated in addiction and reward behavior, lending further support to the concept that obesity, in part, is a centrally-mediated disorder.
Supporting Information
Zdroje
1. CassanoPA
RosnerB
VokonasPS
WeissST
1992 Obesity and body fat distribution in relation to the incidence of non-insulin-dependent diabetes mellitus. A prospective cohort study of men in the normative aging study. Am J Epidemiol 136 1474 1486
2. FolsomAR
KushiLH
AndersonKE
MinkPJ
OlsonJE
2000 Associations of general and abdominal obesity with multiple health outcomes in older women: the Iowa Women's Health Study. Arch Intern Med 160 2117 2128
3. OhlsonLO
LarssonB
SvardsuddK
WelinL
ErikssonH
1985 The influence of body fat distribution on the incidence of diabetes mellitus. 13.5 years of follow-up of the participants in the study of men born in 1913. Diabetes 34 1055 1058
4. WeiM
GaskillSP
HaffnerSM
SternMP
1997 Waist circumference as the best predictor of noninsulin dependent diabetes mellitus (NIDDM) compared to body mass index, waist/hip ratio and other anthropometric measurements in Mexican Americans–a 7-year prospective study. Obes Res 5 16 23
5. RankinenT
ZuberiA
ChagnonYC
WeisnagelSJ
ArgyropoulosG
2006 The human obesity gene map: the 2005 update. Obesity (Silver Spring) 14 529 644
6. LyonHN
HirschhornJN
2005 Genetics of common forms of obesity: a brief overview. Am J Clin Nutr 82 215S 217S
7. ChambersJC
ElliottP
ZabanehD
ZhangW
LiY
2008 Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat Genet 40 716 718
8. FraylingTM
TimpsonNJ
WeedonMN
ZegginiE
FreathyRM
2007 A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316 889 894
9. LoosRJ
LindgrenCM
LiS
WheelerE
ZhaoJH
2008 Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet 40 768 775
10. KringSI
HolstC
ZimmermannE
JessT
BerentzenT
2008 FTO gene associated fatness in relation to body fat distribution and metabolic traits throughout a broad range of fatness. PLoS ONE 3 e2958 doi:10.1371/journal.pone.0002958
11. ThorleifssonG
WaltersGB
GudbjartssonDF
SteinthorsdottirV
SulemP
2008 Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet 41 18 24
12. WillerCJ
SpeliotesEK
LoosRJ
LiS
LindgrenCM
2009 Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet 41 25 34
13. MeyreD
DelplanqueJ
ChevreJC
LecoeurC
LobbensS
2009 Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet 41 157 159
14. ServinB
StephensM
2007 Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet 3 e114 doi:10.1371/journal.pgen.0030114
15. PurcellS
NealeB
Todd-BrownK
ThomasL
FerreiraMA
2007 PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81 559 575
16. O'ConnellJR
2008 Optimizing Measured Genotype Genome-wide Association Analysis for Quantitative Traits in Pedigrees (Abstract program number 2409). ASHG Annual Meeting. Philadelphia, Pennsylvania
17. TeamRDC
2006 R: A language and environment for statistical computing Vienna, Austria R Foundation for Statistical Computing
18. LiY
DingJ
AbecasisGR
2006 Mach 1.0: Rapid Haplotype Reconstruction and Missing Genotype Inference (Abstract program number 2290). ASHG Annual Meeting. New Orleans, Louisiana
19. AulchenkoYS
RipkeS
IsaacsA
van DuijnCM
2007 GenABEL: an R library for genome-wide association analysis. Bioinformatics 23 1294 1296
20. BacanuSA
DevlinB
RoederK
2000 The power of genomic control. Am J Hum Genet 66 1933 1944
21. PriceAL
PattersonNJ
PlengeRM
WeinblattME
ShadickNA
2006 Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38 904 909
22. GordonAGG
QuiX
YakovlevA
2007 Control of the mean number of false discoveries, bonferroni and stability of multiple testing. Annals of Applied Statistics 1 179 190
23. LachmanHM
FannCS
BartzisM
EvgrafovOV
RosenthalRN
2007 Genomewide suggestive linkage of opioid dependence to chromosome 14q. Hum Mol Genet 16 1327 1334
24. KelaiS
MaussionG
NobleF
BoniC
RamozN
2008 Nrxn3 upregulation in the globus pallidus of mice developing cocaine addiction. Neuroreport 19 751 755
25. ClaySW
AllenJ
ParranT
2008 A review of addiction. Postgrad Med 120 E01 07
26. RapakaR
SchnurP
ShurtleffD
2008 Obesity and addiction: common neurological mechanisms and drug development. Physiol Behav 95 2 9
27. BransonR
PotocznaN
KralJG
LentesKU
HoeheMR
2003 Binge eating as a major phenotype of melanocortin 4 receptor gene mutations. N Engl J Med 348 1096 1103
28. GerkenT
GirardCA
TungYC
WebbyCJ
SaudekV
2007 The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 318 1469 1472
29. CecilJE
TavendaleR
WattP
HetheringtonMM
PalmerCN
2008 An obesity-associated FTO gene variant and increased energy intake in children. N Engl J Med 359 2558 2566
30. MyersAJ
GibbsJR
WebsterJA
RohrerK
ZhaoA
2007 A survey of genetic human cortical gene expression. Nat Genet 39 1494 1499
31. DixonAL
LiangL
MoffattMF
ChenW
HeathS
2007 A genome-wide association study of global gene expression. Nat Genet 39 1202 1207
32. SchadtEE
MolonyC
ChudinE
HaoK
YangX
2008 Mapping the genetic architecture of gene expression in human liver. PLoS Biol 6 e107 doi:10.1371/journal.pbio.0060107
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2009 Číslo 6
-
Všechny články tohoto čísla
- Cytoplasmic Streaming in : Disperse the Plug To Increase the Flow?
- The Mysteries of Chromosome Evolution in Gibbons: Methylation Is a Prime Suspect
- Meta-Analysis of 28,141 Individuals Identifies Common Variants within Five New Loci That Influence Uric Acid Concentrations
- The Impact of Divergence Time on the Nature of Population Structure: An Example from Iceland
- Is a Novel Locus for Waist Circumference: A Genome-Wide Association Study from the CHARGE Consortium
- Asymmetric Strand Segregation: Epigenetic Costs of Genetic Fidelity?
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Cytoplasmic Streaming in : Disperse the Plug To Increase the Flow?
- Meta-Analysis of 28,141 Individuals Identifies Common Variants within Five New Loci That Influence Uric Acid Concentrations
- Is a Novel Locus for Waist Circumference: A Genome-Wide Association Study from the CHARGE Consortium
- Asymmetric Strand Segregation: Epigenetic Costs of Genetic Fidelity?
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



