-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaA -Acting Diversification Activator Both Necessary and Sufficient for AID-Mediated Hypermutation
Hypermutation of the immunoglobulin (Ig) genes requires Activation Induced cytidine Deaminase (AID) and transcription, but it remains unclear why other transcribed genes of B cells do not mutate. We describe a reporter transgene crippled by hypermutation when inserted into or near the Ig light chain (IgL) locus of the DT40 B cell line yet stably expressed when inserted into other chromosomal positions. Step-wise deletions of the IgL locus revealed that a sequence extending for 9.8 kilobases downstream of the IgL transcription start site confers the hypermutation activity. This sequence, named DIVAC for diversification activator, efficiently activates hypermutation when inserted at non-Ig loci. The results significantly extend previously reported findings on AID-mediated gene diversification. They show by both deletion and insertion analyses that cis-acting sequences predispose neighboring transcription units to hypermutation.
Published in the journal: . PLoS Genet 5(1): e32767. doi:10.1371/journal.pgen.1000332
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1000332Summary
Hypermutation of the immunoglobulin (Ig) genes requires Activation Induced cytidine Deaminase (AID) and transcription, but it remains unclear why other transcribed genes of B cells do not mutate. We describe a reporter transgene crippled by hypermutation when inserted into or near the Ig light chain (IgL) locus of the DT40 B cell line yet stably expressed when inserted into other chromosomal positions. Step-wise deletions of the IgL locus revealed that a sequence extending for 9.8 kilobases downstream of the IgL transcription start site confers the hypermutation activity. This sequence, named DIVAC for diversification activator, efficiently activates hypermutation when inserted at non-Ig loci. The results significantly extend previously reported findings on AID-mediated gene diversification. They show by both deletion and insertion analyses that cis-acting sequences predispose neighboring transcription units to hypermutation.
Introduction
Vertebrate B cells are able to diversify their rearranged immunoglobulin (Ig) genes by hypermutation, gene conversion and class switch recombination. All three phenomena require expression of Activation Induced cytidine Deaminase (AID, NC_006088) [1]–[3] which most likely initiates Ig gene diversification by deaminating cytidines within the mutating and recombining sequences [4],[5]. A further requisite for hypermutation and switch recombination is the transcription of the Ig genes and the switch regions respectively [6],[7].
Sequence analysis of transcribed non-Ig genes from AID expressing B cells revealed either no or only a low number of mutations compared to Ig genes [8]. A recent study of a large number of expressed genes in B cells found a significantly higher number of mutations in wild-type mice than in AID knock-out mice [9]. However, the mutation rates for the non-Ig genes in AID expressing B cells were still orders of magnitude lower than for the Ig genes. To explain this difference between Ig and non-Ig genes it has been postulated that cis-acting sequences in the Ig loci activate hypermutation possibly by recruiting AID. However, intense efforts did not succeed to unambiguously define these sequences for the murine and human Ig loci [10]. Whereas studies using chimeric reporter genes in transgenic mice indicated that certain Ig enhancers and their surrounding sequences conferred hypermutation activity [11]–[14], deletion of Igκ enhancers in knock-out mice did not prevent hypermutation of the Igκ gene (CAA36032) [15],[16]. At least one murine B cell line [17] and AID expressing fibroblasts [18] mutated transcribed transgenes in the absence of nearby Ig locus sequences, further confounding the issue of whether cis-acting regulatory sequences are needed for hypermutation.
The chicken B cell line DT40 diversifies its rearranged Ig light chain (IgL) gene by gene conversion in the presence of nearby pseudo V (ψV) genes [2] and by hypermutation, if the ψV genes are deleted [19]. Both activities strictly depend on the expression of AID. Consistent with the idea that the absence of homologous gene conversion donors leads to hypermutation, a Green Fluorescent Protein (GFP, AAB08058) transgene is rapidly diversified by mutations when inserted into the rearranged IgL locus [20]. The hypermutation activity of DT40 appeared however to be limited to the IgL locus, because no mutations were found in the highly transcribed Elongation Factor 1 alpha gene (NP_989488) [19]. This was confirmed by a recent study showing that neither the VpreB3 (NC_006102) nor the Carbonic Anhydrase (XP_415218) gene, immediate upstream and downstream neighbors of the IgL locus respectively, showed sequence heterogeneity in DT40 [21].
DT40 has been proposed as a model to study the mechanism of Ig hypermutation [19]. Compared to mice and humans, the chicken IgL locus including the ψV genes is compact spanning only 30 kb. Furthermore, targeted integration of transfected DNA constructs in DT40 allows the introduction of deletions and insertions at defined chromosomal positions. Encouraged by these advantages we decided to search for the elusive cis-acting hypermutation control sequence in the IgL locus of DT40.
Results
A Hypermutation Reporter Based on GFP Expression
We have previously demonstrated that a GFP transgene in DT40 rapidly accumulates mutations, when integrated at the position of the promoter of the rearranged IgL locus [20]. The hypermutation activity depended on AID expression and could be visualized by the appearance of cells displaying decreased green fluorescence due to detrimental GFP mutations. To exploit this phenomenon, we designed a new expression cassette named GFP2 which consisted of the strong RSV promoter followed by the GFP coding region, an internal ribosome entry site (IRES), the blasticidin resistance gene (P19997) and the SV40 polyadenylation signal. GFP2 was incorporated into the targeting construct pIgLGFP2 (Figure 1A) in the opposite transcriptional orientation of the IgL gene to minimize interference between transcriptional and post-transcriptional regulation of the GFP2 transgene and the IgL gene.
Fig. 1. Hypermutation of the GFP2 reporter at various distances from the rearranged IgL locus. 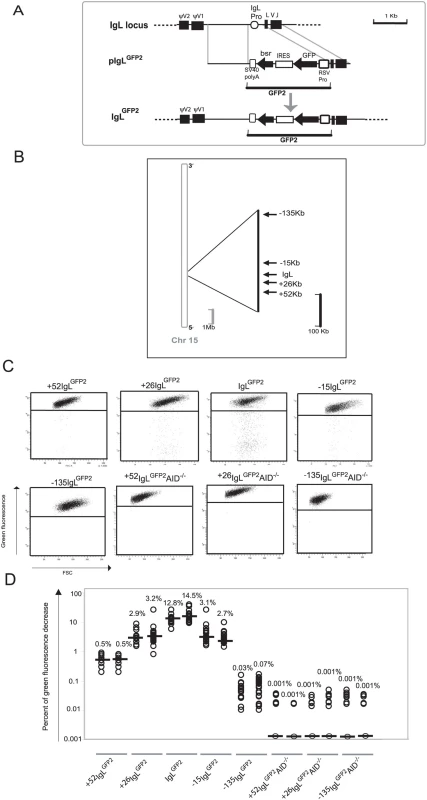
(A) A physical map of the rearranged IgL locus, a targeting construct including the GFP2 reporter and the IgL locus after targeted insertion of the GFP2 reporter. (B) Locations of GFP2 insertion relative to the IgL locus on chromosome 15. The reference point is the insertion site of GFP2 in the IgLGFP2 transfectant. (C) FACS analysis of primary transfectants having integrated the transfected construct targeted. (D) Fluctuation analysis of subclones. Each dot represents the analysis of a single subclone and the median of all subclones from the same primary transfectant is indicated by a bar. Transfection of pIgLGFP2 into the conditionally AID expressing clone AIDR2 yielded a number of transfectants named IgLGFP2 in which targeted integration had substituted the IgL promoter by the GFP2 transgene. Fluorescence activated cell sorting (FACS) analysis of subclones from two independent primary transfectants revealed median values of 12.8% and 14.5% decreased green fluorescence two weeks after subcloning (Figure 1C and 1D). The result confirms our previous study indicating that the GFP2 transgene is mutated at high rate within the rearranged IgL locus and that cell populations with decreased green fluorescence can be used to quantify this hypermutation activity.
Hypermutation in the Vicinity of the IgL Locus
Targeted integration was used to insert the GFP2 reporter at various distances from the IgL locus into chromosome 15 [22] (Figure 1B and Figure S1A). FACS analysis of primary transfectants (Figure 1C and Table S1) and their subclones (Figure 1D) revealed that the medians of decreased green fluorescence fell to about 3% at the +26 kb and the −15 kb positions, to 0.5% at the +52 kb position and to about 0.05% at the −135 kb position. The medians of decreased green fluorescence were only around 0.001% at the +52, +26 and −135 kb positions in the absence of AID (Figure 1C and 1D and Table S1), indicating that the decreased green fluorescence was dependent on AID expression.
Although we did not determine for the GFP2 insertions outside the IgL locus, whether the rearranged or the unrearranged allele was targeted, the results were representative for a large number of independent primary transfectants (Table S1). Thus, hypermutation of the GFP2 reporter was detectable at insertions up to 52 kb away from the IgL locus, but mutations declined with increasing distance and were barely detectable at the −135 kb position.
Since surrounding sequences were unlikely to influence the post-transcriptional processing and translation of GFP2 transcripts, GFP2 transcription should be reflected by the green fluorescence of the cells independent of the transgene insertion site. Even in the case of mutating transgenes, GFP2 transcription levels could be deduced from the average green fluorescence of the major cell populations which most likely expressed the un-mutated GFP sequence. As seen by FACS analysis, the average green fluorescence of the major cell populations varied slightly among the primary transfectants (Figure 1C) most likely reflecting chromosomal position effects. However, the transfectants +52IgLGFP2 and IgLGFP2 differed more than 20 fold in their median fluorescence decreases despite similar green fluorescence of their major cell populations. This strongly suggested that the hypermutation differences among the transfectants reflected the distance of the GFP2 insertion sites to the IgL locus and not variation in GFP2 transcription.
Identification of a Diversification Activator
The results could be explained by the presence of a cis-acting sequence which activated hypermutation in a distance dependent manner. We have named this putative regulatory sequence Diversification Activator (DIVAC) and attempted to map it by combining insertions of the GFP2 reporter with deletions of the IgL locus.
To address the role of the ψV part of the IgL locus, a GFP2 construct (Figure 2A, upper part) was transfected into the clone ψV−AIDR1 [20] in which the entire 20 kb containing the ψV genes had been deleted. The transfectants ψV−IgLGFP2 expressed the GFP2 reporter at the position of the IgL promoter in the absence of the ψV locus.
Fig. 2. GFP2 hypermutation after deletion and reconstitution of the rearranged IgL locus. 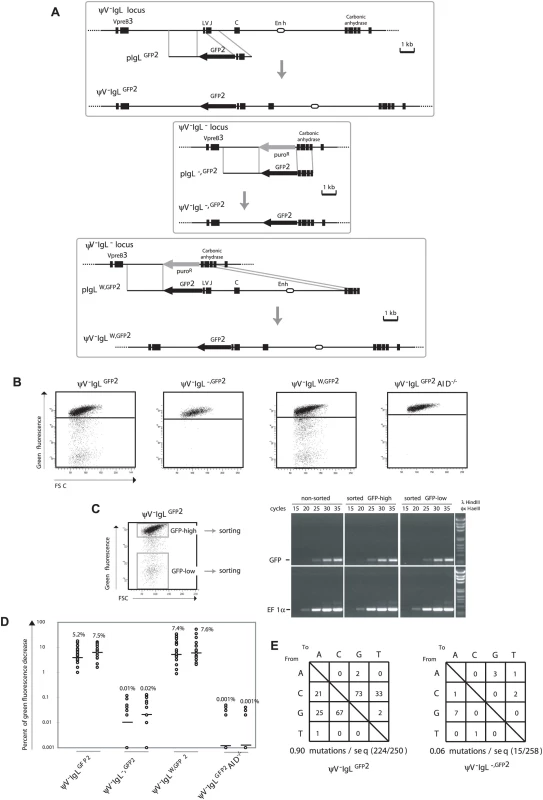
(A) Design of deletions and insertions in the rearranged IgL locus using GFP2 targeting constructs. Upper part: Physical maps of the rearranged IgL locus after the deletion of the ψV locus, a targeting construct including the GFP2 reporter and the locus after targeted integration. Middle part: Physical maps of the deleted rearranged IgL locus, a targeting construct including the GFP2 reporter and the GFP2 insertion site after targeted integration. Lower part: Physical maps of the deleted rearranged IgL locus, a targeting construct including the GFP2 reporter together with the ‘W’ fragment and the locus after targeted integration. (B) FACS analysis of primary transfectants. (C) Sorting of GFP-high and GFP-low cells from a ψV−IgLGFP2 primary transfectant, and semi-quantitative RT-PCR of GFP and EF1α messages from sorted and non-sorted cells. (D) Fluctuation analysis of subclones. (E) The frequencies of particular nucleotide substitutions within the GFP open reading frame of GFP2. FACS analysis of ψV−IgLGFP2 primary transfectants showed sizable populations of cells showing decreased green fluorescence (Figure 2B) indicating that the GFP gene is diversified by hypermutation. To rule out that the decrease of green fluorescence is caused by gene silencing, the populations of high (GFP-high) and low (GFP-low) green fluorescence were sorted from one of the ψV−IgLGFP2 primary transfectants (Figure 2C, left), and GFP mRNA levels was analyzed by semi-quantitative reverse transcription polymerase chain reaction (RT-PCR), (Figure 2C, right). RT-PCR of the EF1α mRNA served as a control. Although GFP-low cells showed on average more than 100-fold lower green fluorescence than GFP-high cells, the levels of GFP mRNA were comparable between sorted GFP-low, GFP-high and non-sorted cells, confirming that the decrease of green fluorescence is not due to silencing of GFP gene expression.
FACS analysis of ψV−IgLGFP2 subclones revealed medians of 5.2% and 7.5% decreased green fluorescence (Figure 2D), only one fold lower than the medians of the ψV positive IgLGFP2 subclones. As the difference between IgLGFP2 and ψV−IgLGFP2 subclones could be due to fluctuation effects or different AID expression levels in the AIDR2 and ψV−AIDR1 precursors, the ψV locus seems to exert little, if any stimulation on the hypermutation activity of the GFP2 reporter.
ψV−IgLGFP2 still contained a 9.8 kb fragment of the rearranged IgL locus extending from the IgL transcription start site until the 3′ end of the carbonic anhydrase gene and referred to in the following as fragment ‘W’. To test the relevance of this fragment, a GFP2 construct was transfected into the clone ψV−IgL− in which the entire rearranged IgL locus had been replaced by the puromycin resistance gene (P42670). The resulting transfectants ψV−IgL−,GFP2 had the puromycin resistance gene replaced by the GFP2 reporter at the position of the deleted IgL locus (Figure 2A, middle part, and Figure 2B). Subclones of ψV−IgL−,GFP2 showed medians of only 0.01% and 0.02% decreased green fluorescence (Figure 2D), more than 100 fold lower than the medians of ψV−IgLGFP2 subclones. This indicated that the ‘W’ fragment, absent in ψV−IgL−,GFP2 but present in ψV−IgLGFP2, was required for hypermutation of the GFP2 transgene. ψV−IgL− cells were then transfected by a construct including the GFP2 transgene and the ‘W’ fragment (Figure 2A, lower part). Subclones of the transfectants ψV−IgLW,GFP2 showed median green fluorescence decreases similar to the medians of ψV−IgLGFP2 subclones (Figure 2D). Thus, the ‘W’ fragment efficiently activates hypermutation after reinsertion into the IgL locus as expected for a true DIVAC sequence.
Controls confirmed that the appearance of cells with decreased green fluorescence reflected hypermutation in the GFP2 gene. As expected, the decrease of green fluorescence in ψV−IgLGFP2 cultures depended on AID, because subclones of the AID negative transfectant ψV−IgLGFP2AID−/− showed only very low medians of 0.001% decreased green fluorescence (Figure 2D). Furthermore, 723 bp of the GFP open reading frame amplified from ψV−IgLGFP2 cells six weeks after subcloning showed an average of 0.9 nucleotide substitutions per sequence (Figure 2E). As the doubling time of the DT40 cell line is about 10 hours, the mutation rate of the GFP gene of ψV−IgLGFP2 was calculated to be 1.3×10−5 mutation/bp/generation, which was similar to the mutation rate of the human hypermutating RAMOS cell line (2.2×10−5 mutation/bp/generation) [23],[24]. The most prevalent mutations were C to G and G to C transversions as previously observed for hypermutation of the IgL VJ segments from ψV deleted DT40 clones [19]. In contrast, only a very low number of nucleotide substitutions, most likely reflecting polymerase chain reaction (PCR) artifacts, were found in the GFP gene of ψV−IgL−,GFP2 cells (Figure 2E).
Fine Mapping of DIVAC
A new series of targeting constructs was transfected into ψV−IgL− to characterize the ‘W’ fragment by step-wise deletions (Figure 3A). FACS analysis of subclones from the different transfectants showed a variable but progressive loss of hypermutation activity when the ‘W’ fragment was shortened from either end (Figure 3C). The 4 kb ‘S’ fragment in the middle of the ‘W’ fragment, which included the previously identified IgL enhancer [25], still produced median green fluorescence decreases of 2.7% and 1.7%. In contrast, the upstream ‘B’ and the downstream ‘P’ fragments on their own produced median green fluorescence decreases of 0.13% and 0.05% respectively, which are low in absolute terms, but clearly above the medians of ψV−IgL−,GFP2. If either one of these fragments was combined with the ‘S’ fragment in the ‘F’ and ‘K’ fragments respectively, the median decreases of green fluorescence were elevated about 3 times. This suggested that the DIVAC of the chicken IgL locus consisted of a central core region and partially redundant flanking regions which contributed to the overall activity. Clearly, more detailed analysis is needed to define the location, the nature and the configuration of the active motifs within the IgL DIVAC.
Fig. 3. Analysis of the ‘W’ fragment deletion series. 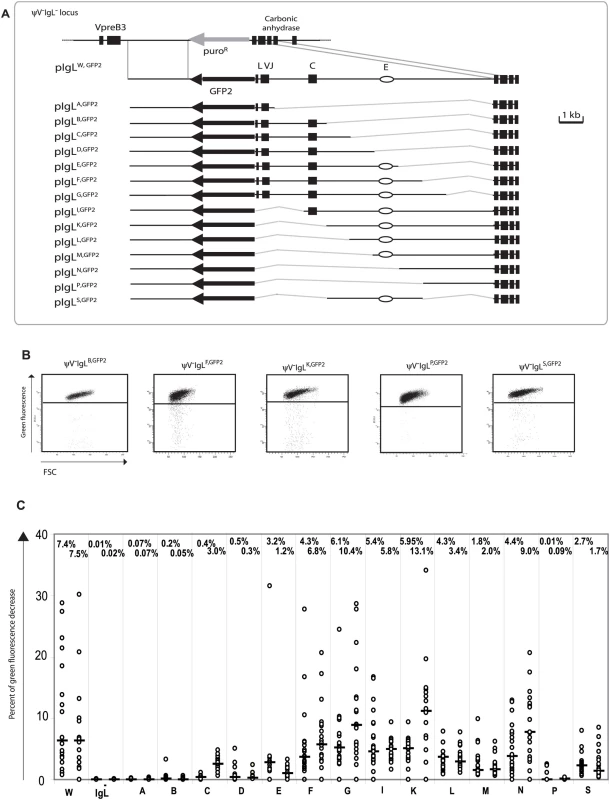
(A) A physical map of the deleted IgL locus and the aligned targeting constructs leading to the insertion of the GFP2 reporter together with parts of the ‘W’ fragment. (B) FACS analysis of representative primary transfectants. (C) Fluctuation analysis of subclones. Only the letter of the reconstituted fragment and not the full name of the transfectants is indicated for clarity. The average green fluorescence in the main population of ψV−IgLW,GFP2 was increased compared to ψV−IgL−,GFP2 (Figure 2B) perhaps due to the additional stimulation of the RSV promoter in GFP2 by the IgL enhancer of the ‘W’ fragment. However, the relatively small decrease of GFP2 transcription seen in ψV−IgL−,GFP2 was unlikely to be responsible for the more than 300 fold reduction of hypermutation. Analysis of the ‘W’ fragment deletions also strongly argued against the possibility that differences in hypermutation were caused by alterations of GFP2 transcription since primary transfectants of all fragments shown in Figure 3B showed similar GFP transcription levels. Similar levels of steady-state GFP2 transcripts were confirmed by RT-PCR (Figure S2).
Hypermutation at non-Ig Loci
To confirm that the GFP2 reporter on its own is stably expressed at non-Ig loci, six loci on five different chromosomes [22] were targeted by transfection of GFP2 constructs into ψV−AIDR1 (Figure 4A and Figure S1B). Neither the primary transfectants (Figure 4B and Table S1B) nor their subclones (Figure 4C) showed high percentages of decreased green fluorescence. Depending on the experiment and the insertion site, the medians of the subclones ranged from 0.02% to 0.22% indicating that the mutation rates of the GFP2 reporter at the chosen loci were 50 to 500 fold lower than at the IgL locus. However, these medians were about 2–10 fold higher than the medians of various subclones from AID negative transfectants (Figures 2D and 5C) confirming a slight increase in the background mutation rates in AID expressing B cells [9].
Fig. 4. Insertions of the GFP2 reporter into non-Ig loci. 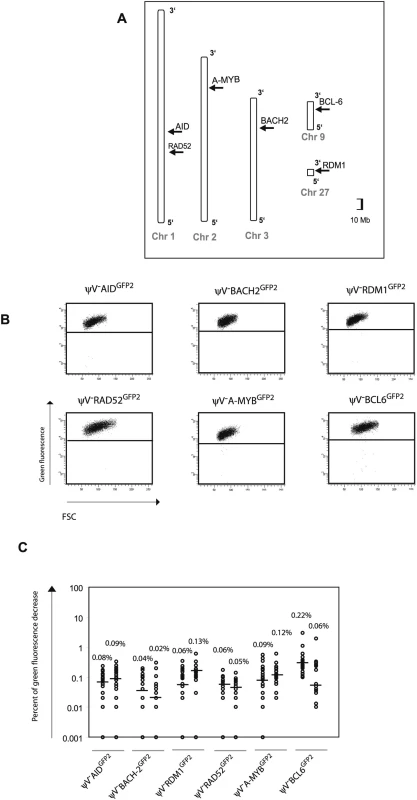
(A) Chromosomal locations of the GFP2 insertions. (B) FACS analysis of primary transfectants. (C) Fluctuation analysis of subclones. GFP2 was then inserted together with the ‘W’ fragment into the respective AID, BACH2 (NC_006090) and RDM1 (BAC02561) loci of ψV−AIDR1 (Figure 5A and 5B). Subclones of the transfectants ψV−AIDW,GFP2, ψV−BACH2W,GFP2 and ψV−RDM1W,GFP2 showed high median decreases of green fluorescence between 4.0% and 9.4% (Figure 5C) similar to the medians for ψV−IgLGFP2 and ψV−IgLW,GFP2 subclones. GFP2 hypermutation was AID dependent, since subclones of the AID negative transfectants ψV−AIDW,GFP2/−, ψV−BACH2W,GFP2/−AID−/− and ψV−RDM1W,GFP2/−AID−/− showed very low medians of decreased green fluorescence in the range of 0.001% to 0.03%. These results demonstrated that the ‘W’ fragment was able to activate AID mediated hypermutation at loci which otherwise did not support hypermutation.
Fig. 5. Hypermutation of the GFP2 reporter at non-Ig loci in the presence of the ‘W’ fragment. 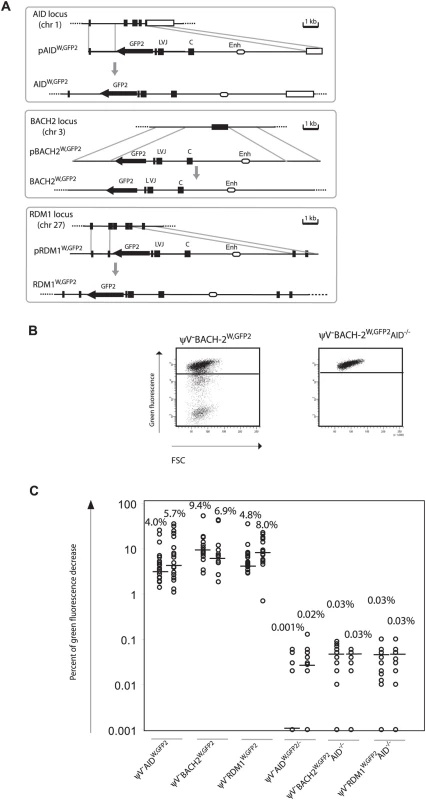
(A) Physical maps of the non-Ig loci, the targeting constructs including the GFP2 reporter together with the ‘W’ fragment and the loci after targeted insertion of the GFP2 reporter. (B) FACS analysis of primary AID positive and AID negative transfectants. (C) Fluctuation analysis of subclones. Discussion
We have identified a cis-acting sequence that is needed for hypermutation at the chicken IgL locus and able to activate hypermutation at other loci upon insertion. The 9.8 kb sequence, named DIVAC for diversification activator, extends from the IgL transcription start site towards the next downstream gene. DIVAC seems to be composed of multiple interacting regions. Whereas a 4 kb core sequence which includes the known IgL enhancer activates hypermutation more than 100 fold above background level, the flanking regions possess less activity on their own, but stimulate hypermutation when combined with the core. Surprisingly, DIVAC can act on either side of the hypermutation reporter and over long distances.
Given the conservation of AID mediated Ig gene diversification during vertebrate evolution, the identification of the chicken IgL DIVAC should also be of relevance for mammals. Searches for cis-acting hypermutation regulatory sequences in transgenic mice showed that regions surrounding the Ig enhancers conferred hypermutation activity [11]–[14]. Intriguingly, the location and functional characteristics of these regions appear to be similar to the core of the chicken IgL DIVAC. In hindsight, the difficulty to unambiguously prove the existence of hypermutation activator sequences may relate to the large size of the murine Ig loci and the fact that DIVACs seem to be composed of multiple interacting regions. As each of the murine Ig loci possesses at least two enhancers at different positions, murine DIVACs may be composed of multiple discontinuous sequences.
Bursal B cells and DT40 in the presence of nearby ψV genes diversify their rearranged IgL loci by gene conversion, suggesting that one of the physiological roles of the IgL DIVAC is activation of gene conversion. This is supported by a recent publication showing that a deletion of the rearranged IgL locus downstream of the C region stopped IgL gene diversification in ψV positive DT40 [26]. It seems also likely that DIVACs play a role for switch recombination which is accompanied by hypermutation of the recombining switch regions [27]. Possibly a dedicated DIVAC near the switch regions activates switch recombination. As the chicken IgL DIVAC can activate hypermutation in both directions over large distances, it is also conceivable that a single DIVAC in the heavy chain loci regulates both hypermutation and switch recombination.
The mechanism of how a cis-regulatory sequence can activate hypermutation in neighboring transcription units remains speculative. Intriguingly, the chicken IgL DIVAC not only includes the IgL enhancer, but also seems to act like an enhancer by activating hypermutation over long distances in upstream or downstream target genes. A plausible hypothesis may be that DIVAC promotes the formation of protein complexes which first bind AID and then hand it over to the neighboring transcription initiation complex. Candidates for proteins involved in building such an AID docking station would be DNA binding factors which recognize sequence motifs within DIVAC. The described experimental system offers unique advantages to test this hypothesis.
Materials and Methods
Targeting Constructs
The GFP2 construct was made by combining the RSV promoter-GFP open reading frame of pHypermut2 [20] with a PCR amplicon including an IRES [19], the blasticidin resistance gene and the SV40 polyadenylation signal [28]. PCR was performed using the primers described in the Table S2. GFP2 was flanked by unique BamHI restriction sites for easy cloning into the targeting vectors.
All targeting constructs except the ones belonging to the series of ‘W’ fragment deletions and reconstitutions were made by cloning the arms sequences into pBluescriptKS+ (Stratagene, CA) and then inserting GFP2 either into unique BamHI or BglII sites as shown in Figure 5A and Figure S1. Targeting of AID [28] and RDM1 [29] have been previously described. PCR amplifications of all target arms were performed using the Expand long template PCR System (Roche, Switzerland), DT40 genomic DNA as template and primers as described in the Table S2.
Since the ‘W’ fragment was difficult to amplify as a single sequence, it was sequentially cloned by combining upstream and downstream PCR amplicons with a 2.2 kb AvrII/SpeI restriction fragment excised from the rearranged IgL targeting construct ‘Construct R’ [30]. The sequence of the AvrII/SpeI restriction fragment is A/T rich and localized between the J segment and the C region. The assembled ‘W’ fragment of 9784 nucleotides was sequenced and deposited into Genbank under the accession number FJ482234. It starts at position −7 relative to the first base of the IgL start codon and corresponds to the chicken genome coordinates chr15 : 8165070–8176699 but lacks the VJ intervening sequence.
Constructs belonging to ‘W’ fragment deletion series were made by cloning GFP2 between the target arms and then inserting the ‘W’ fragment or parts thereof into unique NheI/SpeI sites. A BamHI fragment containing GFP2 and the ‘W’ fragment was incorporated into the AID, BACH2 and RDM1 targeting vectors to test the activity of the ‘W’ fragment in non-Ig loci.
Cell Culture
Cells were cultured in chicken medium (RPMI-1640 or DMEM/F-12 with 10% fetal bovine serum, 1% chicken serum, 2 mM L-glutamine, 0.1 µM β-mercaptoethanol and penicillin/streptomycin) at 41°C with 5% CO2. Transfections were performed by electroporation, using 40 µg of linearized plasmid DNA with a Gene Pulser Xcell (BIO-RAD) at 25 µF and 700 V. Stable transfectants were selected by culturing in 15 µg/ml of blasticidin. Transfectants having integrated the transgenic constructs by targeted integration were identified by PCR using an inside primer from the SV40 polyadenylation signal sequence of GFP2 together with a primer derived from the sequence outside the target arm (Table S2). In case of insertions into the IgL locus, targeted integration into the rearranged allele was verified by amplifying the VJ intervening sequence of the unrearranged locus. The AID reconstituted clone AIDR2 was generated from the AID deleted clone AID−/− [2] by transfection of a construct which targeted an AID cDNA expression cassette into one of the deleted AID loci. The AID negative transfectants were produced by transfecting AID−/− and ψV−AID−/− [19], respectively.
Flow Cytometry
The phenotype of each mutation was determined by FACS analysis of at least two independent targeted transfectants and twenty-four subclones of each. The primary transfectants were analyzed by FACS about three weeks after transfection and the subclones two weeks after subcloning. As the green fluorescence levels in the main populations varied slightly among the transfectants, the gates to separate the main population of green fluorescent cells from cells showing decreased or lost green fluorescence were adapted accordingly. At least 5000 events falling into the live cell gate were collected for each primary transfectant or subclone. Subclones in which more than 50% of the live cell events fell into the gates for decreased or lost green fluorescence were excluded from the analysis as they might represent the expansion of a precursor cell already expressing a mutated GFP2 transgene at the time of subcloning.
GFP Gene Sequencing
To minimize PCR-introduced mutations, Pfu Ultra hotstart polymerase (Stratagene) was used for the amplifications of the GFP open reading frames prior to sequencing. Sequencing and sequence analysis were performed as previously described [19].
RT-PCR
RT-PCR was performed as previously described [2]. Primer pairs used for amplification of the GFP and elongation factor 1α transcripts are shown in Table S2.
Supporting Information
Zdroje
1. MuramatsuM
KinoshitaK
FagarasanS
YamadaS
ShinkaiY
2000 Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102 553 563
2. ArakawaH
HauschildJ
BuersteddeJM
2002 Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science 295 1301 1306
3. HarrisRS
SaleJE
Petersen-MahrtSK
NeubergerMS
2002 AID is essential for immunoglobulin V gene conversion in a cultured B cell line. Curr Biol 12 435 438
4. Di NoiaJ
NeubergerMS
2002 Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature 419 43 48
5. RadaC
Di NoiaJM
NeubergerMS
2004 Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol Cell 16 163 171
6. PetersA
StorbU
1996 Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity 4 57 65
7. ShinkuraR
TianM
SmithM
ChuaK
FujiwaraY
2003 The influence of transcriptional orientation on endogenous switch region function. Nat Immunol 4 435 441
8. ShenHM
PetersA
BaronB
ZhuX
StorbU
1998 Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science 280 1750 1752
9. LiuM
DukeJL
RichterDJ
VinuesaCG
GoodnowCC
2008 Two levels of protection for the B cell genome during somatic hypermutation. Nature 451 841 845
10. OdegardVH
SchatzDG
2006 Targeting of somatic hypermutation. Nat Rev Immunol 6 573 583
11. BetzAG
MilsteinC
González-FernándezA
PannellR
LarsonT
1994 Elements regulating somatic hypermutation of an immunoglobulin kappa gene: critical role for the intron enhancer/matrix attachment region. Cell 77 239 248
12. KlotzEL
StorbU
1996 Somatic hypermutation of a lambda 2 transgene under the control of the lambda enhancer or the heavy chain intron enhancer. J Immunol 157 4458 4463
13. KlixN
JollyCJ
DaviesSL
BrüggemannM
WilliamsGT
1998 Multiple sequences from downstream of the Jκ cluster can combine to recruit somatic hypermutation to a heterologous, upstream mutation domain. Eur J Immunol 28 317 326
14. KongQ
ZhaoL
SubbaiahS
MaizelsN
1998 A λ 3′ enhancer drives active and untemplated somatic hypermutation of a λ1 transgene. J Immunol 161 294 301
15. van der StoepN
GormanJR
AltFW
1998 Reevaluation of 3′Eκ function in stage - and lineage-specific rearrangement and somatic hypermutation. Immunity 8 743 750
16. InlayMA
GaoHH
OdegardVH
LinT
SchatzDG
2006 Roles of the Igκ light chain intronic and 3′ enhancers in Igk somatic hypermutation. J Immunol 177 1146 1151
17. WangCL
HarperRA
WablM
2004 Genome-wide somatic hypermutation. Proc Natl Acad Sci U S A 101 7352 7356
18. YoshikawaK
OkazakiIM
EtoT
KinoshitaK
MuramatsuM
2002 AID enzyme-induced hypermutation in an actively transcribed gene in fibroblasts. Science 296 2033 2036
19. ArakawaH
SaribasakH
BuersteddeJM
2004 Activation-induced cytidine deaminase initiates immunoglobulin gene conversion and hypermutation by a common intermediate. PLoS Biol 2 e179 doi:10.1371/journal.pbio.0020179
20. ArakawaH
KudoH
BatrakV
CaldwellRB
RiegerMA
2008 Protein evolution by hypermutation and selection in the B cell line DT40. Nucleic Acids Res 36 e1
21. GopalAR
FugmannSD
2008 AID-mediated diversification within the IgL locus of chicken DT40 cells is restricted to the transcribed IgL gene. Mol Immunol 45 2062 2068
22. International Chicken Genome Sequencing Consortium 2004 Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432 695 716
23. SaleJ
NeubergerMS
1998 TdT-accessible breaks are scattered over the immunoglobulin V domain in a constitutively hypermutating B cell line. Immunity 9 859 869
24. ZhangW
BardwellPD
WooCJ
PoltoratskyV
ScharffMD
2001 Clonal instability of V region hypermutation in the Ramos Burkitt's lymphoma cell line. Int Immunol 13 1175 1184
25. Bulfone-PausS
Reiners-SchrammL
LausterR
1995 The chicken immunoglobulin lambda light chain gene is transcriptionally controlled by a modularly organized enhancer and an octamer-dependent silencer. Nucleic Acids Res 23 1997 2005
26. KothapalliN
NortonDD
FugmannSD
2008 Cutting edge: a cis-acting DNA element targets AID-mediated sequence diversification to the chicken Ig light chain gene locus. J Immunol 180 2019 2023
27. NagaokaH
MuramatsuM
YamamuraN
KinoshitaK
HonjoT
2002 Activation-induced deaminase (AID)-directed hypermutation in the immunoglobulin Smu region: implication of AID involvement in a common step of class switch recombination and somatic hypermutation. J Exp Med 195 529 534
28. ArakawaH
LodyginD
BuersteddeJM
2001 Mutant loxP vectors for selectable marker recycle and conditional knock-outs. BMC Biotechnol 1 7
29. HamimesS
ArakawaH
StasiakAZ
KierzekAM
HiranoS
2004 RDM1, a novel RNA recognition motif (RRM)-containing protein involved in the cell response to cisplatin in vertebrates. J Biol Chem 280 9225 9235
30. BuersteddeJM
TakedaS
1991 Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell 67 179 188
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2009 Číslo 1
-
Všechny články tohoto čísla
- Order and Disorder during Divergence
- Converging on the Origins of Axonal Ion Channel Clustering
- Mouse Genome-Wide Association Mapping Needs Linkage Analysis to Avoid False-Positive Loci
- Why Is the Correlation between Gene Importance and Gene Evolutionary Rate So Weak?
- A Microhomology-Mediated Break-Induced Replication Model for the Origin of Human Copy Number Variation
- A Novel Role for in the Parallel Evolution of Depigmentation in Independent Populations of the Cavefish
- Copy Number Variation and the Co-Evolution of Primate and Viral Genomes
- Multiple Chromosomal Rearrangements Structured the Ancestral Vertebrate -Bearing Protochromosomes
- A -Acting Diversification Activator Both Necessary and Sufficient for AID-Mediated Hypermutation
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- A -Acting Diversification Activator Both Necessary and Sufficient for AID-Mediated Hypermutation
- Order and Disorder during Divergence
- Mouse Genome-Wide Association Mapping Needs Linkage Analysis to Avoid False-Positive Loci
- Why Is the Correlation between Gene Importance and Gene Evolutionary Rate So Weak?
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



